PROBILITY DISTRIBUTIONS, QUANTILES, CHECKS FOR NORMALITY
Probability Distributions
R has density and distribution functions built-in for about 20 probability
distributions, including those in the following table:
| distribution | function | type |
|---|
| binomial | binom | discrete |
| chi-squared | chisq | continuous |
| F | f | continuous |
| hypergeometric | hyper | discrete |
| normal | norm | continuous |
| Poisson | pois | discrete |
| Student's t | t | continuous |
| uniform | unif | continuous |
By prefixing a "d" to the function name in the table above, you can get
probability density values (pdf). By prefixing a "p", you can get cumulative
probabilities (cdf). By prefixing a "q", you can get quantile values. By
prefixing an "r", you can get random numbers from the distribution. I will
demonstrate using the normal distribution.
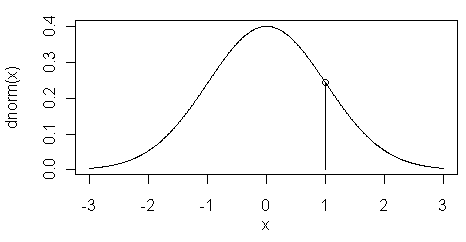
The
dnorm( ) function returns the height of
the normal curve at some value along the x-axis. This is illustrated in the
figure at left. Here the value of
dnorm(1) is shown
by the vertical line at x=1...
> dnorm(1)
[1] 0.2419707
With no options specified, the value of "x" is treated as a standard score or
z-score. To change this, you can specify "mean=" and "sd=" options. In other
words,
dnorm( ) returns the probability
density function or pdf.
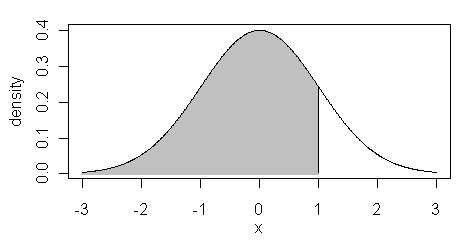
The
pnorm( ) function is the cumulative
density function or cdf. It returns the area below the given value of "x", or
for x=1, the shaded region in the figure at right...
> pnorm(1)
[1] 0.8413447
Once again, the defaults for mean and sd are 0 and 1 respectively. These can be
set to other values as in the case of
dnorm( ). To find the area above the cutoff x-value,
either subtract from 1, or set the "lower.tail=" option to FALSE...
> 1 - pnorm(1)
[1] 0.1586553
> pnorm(1, lower.tail=F)
[1] 0.1586553
So, good news! No more tables!
To get quantiles or "critical values", you can use the
qnorm( ) function as in the following examples...
> qnorm(.95) # p = .05, one-tailed (upper)
[1] 1.644854
> qnorm(c(.025,.975)) # p = .05, two-tailed
[1] -1.959964 1.959964
> qnorm(seq(.1,.9,.1)) # deciles from the unit normal dist.
[1] -1.2815516 -0.8416212 -0.5244005 -0.2533471 0.0000000 0.2533471 0.5244005
[8] 0.8416212 1.2815516
Once again, there are "mean=" and "sd=" options.
To use these functions with other distributions, more parameters may need to
be given. Here are some examples...
> pt(2.101, df=8) # area below t = 2.101, df = 8
[1] 0.9655848
> qchisq(.95, df=1) # critical value of chi square, df = 1
[1] 3.841459
> qf(c(.025,.975), df1=3, df2=12)
[1] 0.06975178 4.47418481
> dbinom(60, size=100, prob=.5) # a discrete binomial probability
[1] 0.01084387
The help pages for these functions will give the necessary details.
Random numbers are generated from a given distribution like this...
> runif(9) # 9 uniformly distributed random nos.
[1] 0.01961714 0.62086249 0.64193142 0.99583719 0.06294405 0.94324289 0.88233387
[8] 0.11851026 0.60300929
> rnorm(9) # 9 normally distributed random nos.
[1] -0.95186711 0.09650050 -0.37148202 0.56453509 -0.44124876 -0.43263580
[7] -0.46909466 1.38590806 -0.06632486
> rt(9, df=10) # 9 t-distributed random nos.
[1] -1.538466123 -0.249067184 -0.324245905 -0.009964799 0.143282490
[6] 0.619253016 0.247399305 0.691629869 -0.177196453
One again, I refer you to the help pages for all the gory details.
Empirical Quantiles
Suppose you want quartiles or deciles or percentiles or whatever from a
sample or empirical distribution. The appropriate function is
quantile( ). From the help page for this function,
the syntax is...
quantile(x, probs = seq(0, 1, 0.25), na.rm = FALSE,
names = TRUE, type = 7, ...)
This says "enter a vector, x, of data values, or the name of such a vector, and
I will return quantiles for positions 0, .25, .5, .75, and 1 (in other words,
quartiles along with the min and max values), without removing missing values
(and if missing values exist the function will fail and return an error
message), I'll give each of the returned values a name, and I will use method 7
(of 9) to do the calculations." Let's see this happen using the built-in data
set "rivers"...
> quantile(rivers)
0% 25% 50% 75% 100%
135 310 425 680 3710
Compare this to what you get with a summary...
> summary(rivers)
Min. 1st Qu. Median Mean 3rd Qu. Max.
135.0 310.0 425.0 591.2 680.0 3710.0
So what's the point? The
quantile( ) function
is much more versatile because you can change the default "probs=" values...
> quantile(rivers, probs=seq(.2,.8,.2)) # quintiles
20% 40% 60% 80%
291 375 505 735
> quantile(rivers, probs=seq(.1,.9,.1)) # deciles
10% 20% 30% 40% 50% 60% 70% 80% 90%
255 291 330 375 425 505 610 735 1054
> quantile(rivers, probs=.55) # 55th percentile
55%
460
> quantile(rivers, probs=c(.05,.95)) # and so on
5% 95%
230 1450
And then there is the "type=" option. It turns out there is some disagreement
among different sources as to just how quantiles should be calculated from an
empirical distribution. R doesn't take sides. It gives you nine different
methods! Pick the one you like best by setting the "type=" option to a number
between 1 and 9. Here are some details (and more are available on the help
page): type=2 will give the results most people are taught to calculate in an
intro stats course, type=3 is the SAS definition, type=6 is the Minitab and
SPSS definition, type=7 is the default and the S definition and seems to work
well when the variable is continuous.
Checks For Normality
Parametric procedures like the t-test, F-test (ANOVA), and Pearson r assume
the data are distributed normally. There are several ways to check this
assumption.
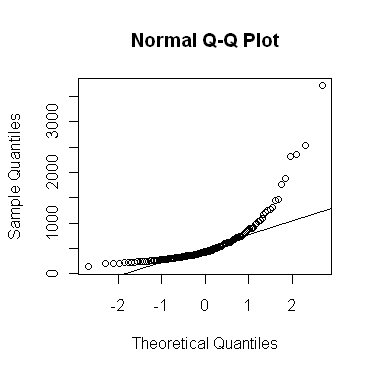
The
qqnorm( ) function allows a graphical
evaluation...
> qqnorm(rivers)
If the values in the vector are normally distributed, the points on the plot
will fall (more or less) along a straight line. This line can be plotted on the
graph like this...
> qqline(rivers)
As you can see, the "rivers" vector is strongly skewed, as indicated by the
bowing of the points up away from the expected straight line. The very long
upper tail (strong positive skew) in this distribution could also have been
visualized using...
> plot(density(rivers))
...the output of which is not shown here. You can see this in the QQ plot as
well by the fact that the higher sample values are much too large to be from a
theoretical normal distribution. The lower tail of the distribution appears to
be a bit short.
Statistical tests for normality are also available. Perhaps the best known
of these is the Shapiro-Wilk test...
> shapiro.test(rivers)
Shapiro-Wilk normality test
data: rivers
W = 0.6666, p-value < 2.2e-16
I believe we can safely reject the null hypothesis of normality here! Here's a
question for all you stat students out there: how often should the following
result in a rejection of the null hypothesis if our random number generator is
worth its salt?
> shapiro.test(rnorm(100))
Shapiro-Wilk normality test
data: rnorm(100)
W = 0.9894, p-value = 0.6192
This question will be on the exam!
Another test that can be used here is the Kolmogorov-Smirnov test...
> ks.test(rivers, "pnorm", alternative="two.sided")
One-sample Kolmogorov-Smirnov test
data: rivers
D = 1, p-value < 2.2e-16
alternative hypothesis: two-sided
Warning message:
In ks.test(rivers, "pnorm", alternative = "two.sided") :
cannot compute correct p-values with ties
But it gets upset when there are ties in the data. Inside the function, the
value "pnorm" tells the test to compare the empirical cumulative density
function of "rivers" to the cumulative density function of a normal
distribution. The null hypothesis says the two will match. Clearly they do not,
so the null hypothesis is rejected. We conclude once again, and for the last
time in this tutorial, that "rivers" is not normally distributed.



 The
The  The
The  The
The 
 Rob Tougher
Rob Tougher